High-throughput methodologies
The advancement of next-generation sequencing (NGS) technology has opened up many exciting opportunities for high-throughput methodology development. One of my research focuses is to develop NGS-based methodologies to study viruses.Monitoring viral quasispecies by next-generation sequencing
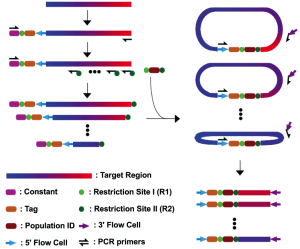 Trade-offs between throughput, read length, and error rates in NGS limit certain applications such as monitoring viral quasispecies. I have developed a molecular-based tag linkage method that allows assemblage of short sequence reads into long DNA fragments. It enables haplotype phasing with high accuracy and sensitivity to interrogate individual viral sequences in a quasispecies. In addition to the molecular methods, I have also involved in the development of computational algorithms for reconstructing quasispecies from Illumina short-read data and for correcting sequencing errors from PacBio long-read data. These approaches can improve monitoring of the genetic architecture and evolution dynamics in any quasispecies population.
Trade-offs between throughput, read length, and error rates in NGS limit certain applications such as monitoring viral quasispecies. I have developed a molecular-based tag linkage method that allows assemblage of short sequence reads into long DNA fragments. It enables haplotype phasing with high accuracy and sensitivity to interrogate individual viral sequences in a quasispecies. In addition to the molecular methods, I have also involved in the development of computational algorithms for reconstructing quasispecies from Illumina short-read data and for correcting sequencing errors from PacBio long-read data. These approaches can improve monitoring of the genetic architecture and evolution dynamics in any quasispecies population.References: Wu et al. 2014a, Mangul et al. 2014, Artyomenko et al. 2017
Searching for essential genetic elements by high-throughput genetics
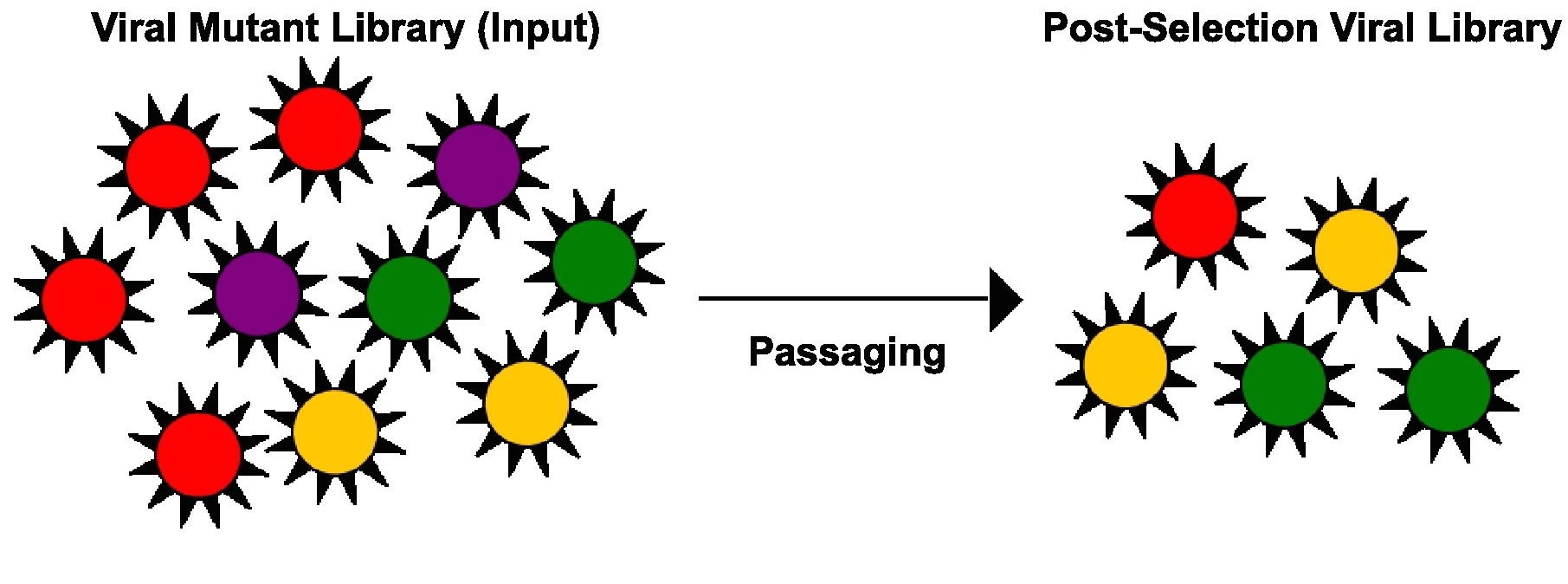 High-throughput genetics aims to study the phenotypic outcome of multiple mutants simultaneously. One strategy is to employ a mutant library consisting of a large number of mutants and to monitor the enrichment or diminishment, hence the phenotypic outcome, of each mutant. By studying all possible mutations on the viral genome in parallel, regions that cannot tolerate mutation will be identified. Such data will be informative for antiviral agent development.
High-throughput genetics aims to study the phenotypic outcome of multiple mutants simultaneously. One strategy is to employ a mutant library consisting of a large number of mutants and to monitor the enrichment or diminishment, hence the phenotypic outcome, of each mutant. By studying all possible mutations on the viral genome in parallel, regions that cannot tolerate mutation will be identified. Such data will be informative for antiviral agent development.References: Wu et al. 2014b, Wu et al. 2015, Wu et al. 2016